Discovering New Bacteria Relevant to Public Health
When patients with infectious diseases seek treatment, they give samples that go to medical laboratories. Labs analyze the samples for pathogens – microorganisms like bacteria, viruses, and fungi that are known to cause diseases. Sometimes the labs discover microorganisms that they can’t identify with the tools they have in their labs.
Medical labs then send the microorganisms to reference laboratories within larger organizations like the Minnesota Department of Health and the Mayo Clinic. If the experts at reference labs cannot identify a microorganism, it remains a mystery until the opportunity arises to fully analyze it and possibly get it named a new species.
 Only after a microorganism is characterized and named can researchers across the world make connections between microorganisms causing disease in more than one patient. Without a name and microorganism characteristics, it can remain a mystery as to what disease such organisms may cause. If a microorganism is named, and a lab discovers it in another patient, doctors can draw on research about that microorganism to plan treatments.
Only after a microorganism is characterized and named can researchers across the world make connections between microorganisms causing disease in more than one patient. Without a name and microorganism characteristics, it can remain a mystery as to what disease such organisms may cause. If a microorganism is named, and a lab discovers it in another patient, doctors can draw on research about that microorganism to plan treatments.
With funding from the Pathogen Genomics Centers of Excellence (PGCoE), the Mayo Clinic launched a program to discover and name at least eight new bacteria that are relevant to public health. In January 2025, Mayo Clinic researchers led by Professor Robin Patel announced their first discovery, the bacteria Corynebacterium mayonis.
Corynebacterium is an established genus of bacteria that includes known pathogens. Prof. Patel et al. chose the species name “mayonis” to pay tribute to the Mayo Clinic. The discovery was published in the article “Corynebacterium mayonis sp. nov. isolated from a human blood culture,” which appeared in the leading journal for announcing new species, the International Journal of Systematic and Evolutionary Microbiology.
The Challenges of Naming a New Species
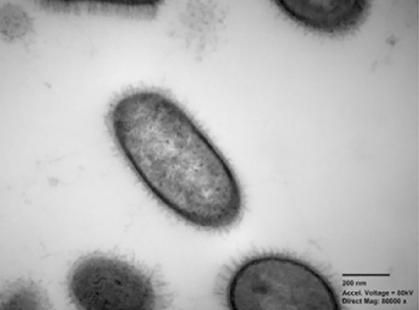 Naming a new species of bacteria involves a lot more than coming up with a good name. Extensive research and characterization of the microorganism is necessary so that medical professionals across the world can study the bacteria with full confidence that they are all gathering knowledge about the same organism.
Naming a new species of bacteria involves a lot more than coming up with a good name. Extensive research and characterization of the microorganism is necessary so that medical professionals across the world can study the bacteria with full confidence that they are all gathering knowledge about the same organism.
Professor Patel and her group, including research technologist Christina Koscianski, began by compiling an extremely detailed description of their bacteria. They used whole-genome sequencing (WGS) to assemble a full profile of its genome. They also studied how the bacteria grows, what it looks like, and other observable characteristics that are collectively known as a “phenotype.”
With the first species named, Professor Patel and her team have now established a pathway that will make identifying future species a smoother process. They have many more candidates for novel species in their laboratory and will continue work on naming new species for as long as the PGCoE funding allows.

 Only after a microorganism is characterized and named can researchers across the world make connections between microorganisms causing disease in more than one patient. Without a name and microorganism characteristics, it can remain a mystery as to what disease such organisms may cause. If a microorganism is named, and a lab discovers it in another patient, doctors can draw on research about that microorganism to plan treatments.
Only after a microorganism is characterized and named can researchers across the world make connections between microorganisms causing disease in more than one patient. Without a name and microorganism characteristics, it can remain a mystery as to what disease such organisms may cause. If a microorganism is named, and a lab discovers it in another patient, doctors can draw on research about that microorganism to plan treatments. Naming a new species of bacteria involves a lot more than coming up with a good name. Extensive research and characterization of the microorganism is necessary so that medical professionals across the world can study the bacteria with full confidence that they are all gathering knowledge about the same organism.
Naming a new species of bacteria involves a lot more than coming up with a good name. Extensive research and characterization of the microorganism is necessary so that medical professionals across the world can study the bacteria with full confidence that they are all gathering knowledge about the same organism.